John I. Pitt, Lene Lange, Alastair E. Lacey, Daniel Vuong, David J. Midgley, Paul Greenfield, Mark I. Bradbury, Ernest Lacey, Peter K. Busk, Bo Pilgaard, Yit-Heng Chooi, Andrew M. Piggott
PLoS ONE 12(4), e0170254.
Publication Date: April 5, 2017
https://doi.org/10.1371/journal.pone.0170254
Abstract:
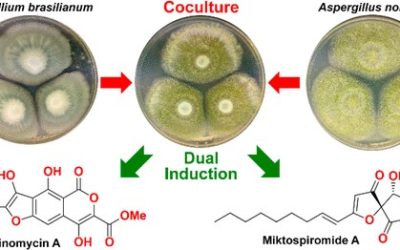
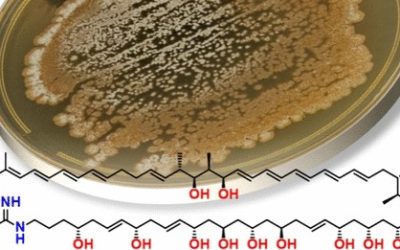
Aspergillus hancockii sp. nov., classified in Aspergillus subgenus Circumdati section Flavi, was originally isolated from soil in peanut fields near Kumbia, in the South Burnett region of southeast Queensland, Australia, and has since been found occasionally from other substrates and locations in southeast Australia. It is phylogenetically and phenotypically related most closely to A. leporis States and M. Chr., but differs in conidial colour, other minor features and particularly in metabolite profile. When cultivated on rice as an optimal substrate, A. hancockii produced an extensive array of 69 secondary metabolites. Eleven of the 15 most abundant secondary metabolites, constituting 90% of the total area under the curve of the HPLC trace of the crude extract, were novel. The genome of A. hancockii, approximately 40 Mbp, was sequenced and mined for genes encoding carbohydrate degrading enzymes identified the presence of more than 370 genes in 114 gene clusters, demonstrating that A. hancockii has the capacity to degrade cellulose, hemicellulose, lignin, pectin, starch, chitin, cutin and fructan as nutrient sources. Like most Aspergillus species, A. hancockii exhibited a diverse secondary metabolite gene profile, encoding 26 polyketide synthase, 16 nonribosomal peptide synthase and 15 nonribosomal peptide synthase-like enzymes.