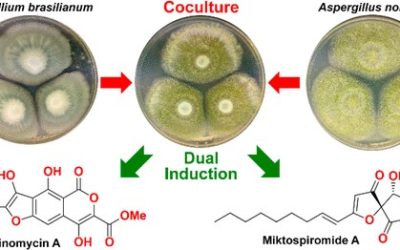
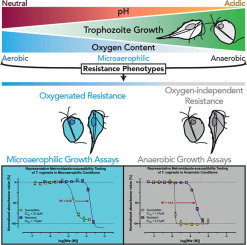
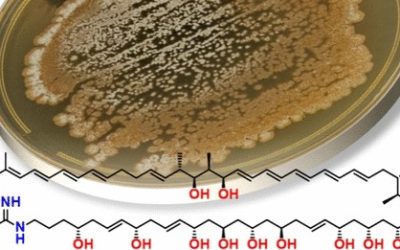
Samantha J. Emery, Ernest Lacey, Paul A. Haynes
Data in Brief 2015, 5, 23–27.
Publication Date: August 19, 2015
https://doi.org/10.1016/j.dib.2015.08.003
Abstract:
Eight Assemblage A strains from the protozoan parasite Giardia duodenalis were analysed using label-free quantitative shotgun proteomics, to evaluate inter- and intra-assemblage variation and complement available genetic and transcriptomic data. Isolates were grown in biological triplicate in axenic culture, and protein extracts were subjected to in-solution digest and online fractionation using Gas Phase Fractionation (GPF). Recent reclassification of genome databases for subassemblages was evaluated for database-dependent loss of information, and proteome composition of different isolates was analysed for biologically relevant assemblage-independent variation. The data from this study are related to the research article “Quantitative proteomics analysis of Giardia duodenalis Assemblage A – a baseline for host, assemblage and isolate variation” published in Proteomics (Emery et al., 2015 [1]).