Amr A. Arishi, Zhuo Shang, Ernest Lacey, Andrew Crombie, Daniel Vuong, Hang Li, Joe Bracegirdle, Peter Turnerg, William Lewis, Gavin R. Flematti, Andrew M. Piggott and Yit-Heng Chooi
Chem. Sci., 2024
Publication Date: January 29, 2024
Abstract:
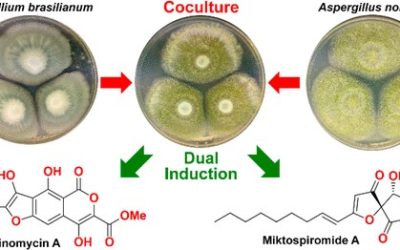
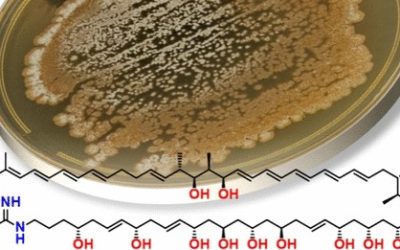
Luteodienoside A is a novel glycosylated polyketide produced by the Australian fungus Aspergillus luteorubrus MST-FP2246, consisting of an unusual 1-O-β-D-glucopyranosyl-myo-inositol (glucinol) ester of 3-hydroxy-2,2,4-trimethylocta-4,6-dienoic acid. Mining the genome of A. luteorubrus identified a putative gene cluster for luteodienoside A biosynthesis (ltb), harbouring a highly reducing polyketide synthase (HR-PKS, LtbA) fused at its C-terminus to a carnitine O-acyltransferase (cAT) domain. Heterologous pathway reconstitution in Aspergillus nidulans, substrate feeding assays and gene truncation confirmed the identity of the ltb cluster and demonstrated that the cAT domain is essential for offloading luteodienoside A from the upstream HR-PKS. Unlike previously characterised cAT domains, the LtbA cAT domain uses glucinol as an offloading substrate to release the product from the HR-PKS. Furthermore, the PKS methyltransferase (MT) domain is capable of catalysing gem-dimethylation of the 3-hydroxy-2,2,4-trimethylocta-4,6-dienoic acid intermediate, without requiring reversible product release and recapture by the cAT domain. This study expands the repertoire of polyketide modifications known to be catalysed by cAT domains and highlights the potential of mining fungal genomes for this subclass of fungal PKSs to discover new structurally diverse secondary metabolites.